For those wanting to watch Netflix and Hulu on an HDTV check out this review of streaming media players, including: Roku, Boxee, and Apple TV.
Showing posts with label scitech. Show all posts
Showing posts with label scitech. Show all posts
Tuesday, February 04, 2014
Review of 5 HDTV Indoor Antennas: Mohu Leaf, Terk FDTV2A, Antennas Direct Clearstream 2, RCA 1450B and Rabbit Ears
For those wanting to watch Netflix and Hulu on an HDTV check out this review of streaming media players, including: Roku, Boxee, and Apple TV.
Thursday, August 30, 2012
Installing and Setting Up HaxeFlixel on Windows
HaxeFlixel is a Flixel port for Haxe the "universal" programming language and the NME application framework to support applications on iOS, Android, webOS, BlackBerry, Windows, Mac, Linux and Flash Player without requiring code changes for each target; Flixel is a popular library for developing 2D Flash games. This tutorial will teach you how to install Haxe, NME, HaxeFlixel, their dependencies, and try out one of the HaxeFlixel demos using Flash.
Monday, February 20, 2012
Cleaning Data with Google Refine and Visualization with Fusion Tables
Introduction
Google Refine is a powerful faceted browser for cleaning up data from large data sets that might contain errors and connecting that data to external databases (known as reconciliation). As an example this article will go through the steps of creating a data visualization using Google Maps to show the locations of universities for a list of graduate students, below is the final product. This process involves using many of the main data cleaning and mining features of Google Refine.
Data Clean Up
Once Google Refine is downloaded and unzipped, it runs in a browser at http://127.0.0.1:3333 and does not require installation. Google Refine can handle large data sets and the data can be imported into Google Refine using many common formats, like delimited data (e.g. comma or tab-delimited), Excel, JSON, and XML. To create the project, the list of students and their universities were imported as a tab-delimited file on the "Create Project" screen; Google Refine automatically determined that the data was tab-delimited, and it assumed that the first row included column names. Google Refine provides a preview window where users can change import settings, such as delimiters, selecting column headings, etc.You can also do some data clean up here by ignore comment rows that might be at the top of the data file.
Each column in Google Refine has a drop-down menu for filtering (known as faceting), sorting, and reconciling. It should be noted that during this entire process, it is easy to undo any changes using the "Undo / Redo" tab in the left hand sidebar. Options for reconciling are in the "Reconcile" menu shown below and the first step is to select "Start reconciling..." in the submenu.
This brings up a new window where pre-existing reconciliation services can be selected or new services can be added. Google Refine by default includes two Freebase-based services that are useful for more powerful data mining; these were used to get the latitude and longitude locations for the universities. Google Refine, again, tries to automatically determine what type of data is in the column; in this case it guessed correctly, and the first option is "College/University" with options like "Educational Institution", also, being in the list.
Once the reconciliation is done, several changes happened in the interface. Under the "Graduate University" column name, a green bar appeared to give a visual cue as to how many universities were matched. Within the column, universities that were correctly matched were highlighted in blue. Those that were not matched correctly had possible options below them with a check mark box to select the option; there was also a double check mark box that repeated the option for all identical entries.
Two new boxes then appeared in the sidebar; these are the facets (filters). The judgment box showed counts for the students who had blank entries, had their universities matched, or where there was no match. The second box was a histogram that visualizes the scores for the matching. Clicking on the "(blank)" entry in the judgment box causes only the rows with a blank entry to be shown; multiple selections can be made using the "include" option that appears on mouse over.
For the initial data cleanup, the students with blank entries were deleted by first clicking on the "(blank)" entry. Then the deleting was done in the data display window by clicking on the "Remove all matching rows" option in the "Edit rows" submenu of the "All" column drop-down menu. In the example case, this left about 30 rows not being reconciled because the university name contained a secondary affiliation acronym. These acronyms needed to be removed and reconciliation needed to be redone. There are two ways of editing the cell values: 1) Mousing over a cell in the data display brings up an edit menu to change the cell value (shown below); all cells with matching contents can be changed and 2) a transformation can be done on all the cells at once.
This example used the second option. The acronyms always appear in the form "acronym / university name", so a single transformation was done on the cells. In the "Graduate University" drop-down menu there is an the option to "Edit cells" with the submenu option "Transform..." that brings up the window shown below where the Google Refine Expression Language (GREL) is used to transform the cell values; Clojure and Jython-based expressions can also be used if you need to clean up data in a more advanced manner. The window has two main sections: 1) a text area to input an expression and 2) a preview section to see how the column will be transformed; this section also gives tabs to see past expressions.
By default, the expression is "value"; "value" is the current value of the cell, and expressions are applied to this value. To select out the university name, the "match" function was used. The match function works by using regular expressions to match some part of the string; all the accessible GREL functions can be found here. Below is the expression that will pull out just the university name.
Retrieving Latitude and Longitude
With the universities cleaned up, the next step was to retrieve the latitudes and longitudes by adding columns with this information from Freebase.
Selecting the "Add columns from Freebase ..." option in the screenshot above brings up a window where various pieces of information can be pulled back; for this example, first, GeoLocation >> Longitude was selected, and then, this was repeated with Latitude. Some of the entries were missing, so, for simplicity, these rows were removed. To do this, the first step was to create facet to identify the blank entries. Facets are created from the column drop-down menu "Facet", then "Text Facet" for either the latitude or longitude columns (this assumes that the absence of one means the absence of the other). Creating this facet will create a new box in the sidebar. The last option is the "(blank)" option used to select only the blank entries and these rows were deleted using the option from the "All" column, as before. Lastly, at the top of the interface is an option to export the results into several formats; "Tab-separated value" was selected because it is easily imported into other software.
Generating a Google Map
Visual representation of data can be very useful when dealing with large data sets; here is a technique for visualizing data on Google Maps. The university locations were placed onto a Google Map using Google Fusion Tables; this is a beta feature of Google Docs. Creating a new table brings up an import dialog. Once the import is complete, Fusion Tables will have a Visualize menu at the top with a visualize as Map option; this is shown below. By selecting this option, Fusion Tables should then be able to automatically take the latitude and longitude columns and render them on a Google Map for a nice visualization; this is the first figure of the article. The map can then be exported as KML or embedded in another webpage.
Google Refine is a powerful faceted browser for cleaning up data from large data sets that might contain errors and connecting that data to external databases (known as reconciliation). As an example this article will go through the steps of creating a data visualization using Google Maps to show the locations of universities for a list of graduate students, below is the final product. This process involves using many of the main data cleaning and mining features of Google Refine.
Once Google Refine is downloaded and unzipped, it runs in a browser at http://127.0.0.1:3333 and does not require installation. Google Refine can handle large data sets and the data can be imported into Google Refine using many common formats, like delimited data (e.g. comma or tab-delimited), Excel, JSON, and XML. To create the project, the list of students and their universities were imported as a tab-delimited file on the "Create Project" screen; Google Refine automatically determined that the data was tab-delimited, and it assumed that the first row included column names. Google Refine provides a preview window where users can change import settings, such as delimiters, selecting column headings, etc.You can also do some data clean up here by ignore comment rows that might be at the top of the data file.
Each column in Google Refine has a drop-down menu for filtering (known as faceting), sorting, and reconciling. It should be noted that during this entire process, it is easy to undo any changes using the "Undo / Redo" tab in the left hand sidebar. Options for reconciling are in the "Reconcile" menu shown below and the first step is to select "Start reconciling..." in the submenu.
This brings up a new window where pre-existing reconciliation services can be selected or new services can be added. Google Refine by default includes two Freebase-based services that are useful for more powerful data mining; these were used to get the latitude and longitude locations for the universities. Google Refine, again, tries to automatically determine what type of data is in the column; in this case it guessed correctly, and the first option is "College/University" with options like "Educational Institution", also, being in the list.
Once the reconciliation is done, several changes happened in the interface. Under the "Graduate University" column name, a green bar appeared to give a visual cue as to how many universities were matched. Within the column, universities that were correctly matched were highlighted in blue. Those that were not matched correctly had possible options below them with a check mark box to select the option; there was also a double check mark box that repeated the option for all identical entries.
Two new boxes then appeared in the sidebar; these are the facets (filters). The judgment box showed counts for the students who had blank entries, had their universities matched, or where there was no match. The second box was a histogram that visualizes the scores for the matching. Clicking on the "(blank)" entry in the judgment box causes only the rows with a blank entry to be shown; multiple selections can be made using the "include" option that appears on mouse over.
For the initial data cleanup, the students with blank entries were deleted by first clicking on the "(blank)" entry. Then the deleting was done in the data display window by clicking on the "Remove all matching rows" option in the "Edit rows" submenu of the "All" column drop-down menu. In the example case, this left about 30 rows not being reconciled because the university name contained a secondary affiliation acronym. These acronyms needed to be removed and reconciliation needed to be redone. There are two ways of editing the cell values: 1) Mousing over a cell in the data display brings up an edit menu to change the cell value (shown below); all cells with matching contents can be changed and 2) a transformation can be done on all the cells at once.
This example used the second option. The acronyms always appear in the form "acronym / university name", so a single transformation was done on the cells. In the "Graduate University" drop-down menu there is an the option to "Edit cells" with the submenu option "Transform..." that brings up the window shown below where the Google Refine Expression Language (GREL) is used to transform the cell values; Clojure and Jython-based expressions can also be used if you need to clean up data in a more advanced manner. The window has two main sections: 1) a text area to input an expression and 2) a preview section to see how the column will be transformed; this section also gives tabs to see past expressions.
By default, the expression is "value"; "value" is the current value of the cell, and expressions are applied to this value. To select out the university name, the "match" function was used. The match function works by using regular expressions to match some part of the string; all the accessible GREL functions can be found here. Below is the expression that will pull out just the university name.
value.match(/.*\/\s(.*)/)[0]Parentheses surround the part of the cell value to be retrieved. Multiple selections can be made, which are returned as an array. Here, only one selection is made so it is array entry "0". After the transformation, the reconciliation was repeated, which brought down the unmatched count to 5. Another way to reconcile is to do a manual search; this is the option that appears below the check mark options and was used to eliminate the remaining few erroneous entries.
Retrieving Latitude and Longitude
With the universities cleaned up, the next step was to retrieve the latitudes and longitudes by adding columns with this information from Freebase.
Selecting the "Add columns from Freebase ..." option in the screenshot above brings up a window where various pieces of information can be pulled back; for this example, first, GeoLocation >> Longitude was selected, and then, this was repeated with Latitude. Some of the entries were missing, so, for simplicity, these rows were removed. To do this, the first step was to create facet to identify the blank entries. Facets are created from the column drop-down menu "Facet", then "Text Facet" for either the latitude or longitude columns (this assumes that the absence of one means the absence of the other). Creating this facet will create a new box in the sidebar. The last option is the "(blank)" option used to select only the blank entries and these rows were deleted using the option from the "All" column, as before. Lastly, at the top of the interface is an option to export the results into several formats; "Tab-separated value" was selected because it is easily imported into other software.
Generating a Google Map
Visual representation of data can be very useful when dealing with large data sets; here is a technique for visualizing data on Google Maps. The university locations were placed onto a Google Map using Google Fusion Tables; this is a beta feature of Google Docs. Creating a new table brings up an import dialog. Once the import is complete, Fusion Tables will have a Visualize menu at the top with a visualize as Map option; this is shown below. By selecting this option, Fusion Tables should then be able to automatically take the latitude and longitude columns and render them on a Google Map for a nice visualization; this is the first figure of the article. The map can then be exported as KML or embedded in another webpage.
Tuesday, March 29, 2011
Run Tiddlywiki on Android Using Firefox
Firefox for Android is capable of saving Tiddlywiki files stored locally on an Android phone. To access the wiki just point to the wiki file on the phone in the address bar. For instance, if the Tiddlywiki file is called wiki.html and stored on your SD card, the URL would be "file:///mnt/sdcard/wiki.html" without the quotes.
Sunday, February 28, 2010
Scientific Bonus Points for Database Contributions
The most common method for assessing the performance of researchers is by looking at a researcher's publication record and the journal impact factor of those the publications. A more recent development in quantifying research output is the h-index by Jorge Hirsch in 2005 that looks at scientific output also as a measure of publication citations. Bornmann and Daniel outline the methodology of the h-index and various concerns about using the metric in their 2008 publication. For those curious about their own h-index value, here is an h-index calculator built using Google Scholar. But what else do researchers do that could be seen as "scientific output" and how could these activities be measured? As a computational biologist, I have a specific interest in online databases, which often serve as the main sources of data for projects in my field, and ways that researchers could be encouraged to contribute to them.
Jesse Schell, a Carnegie Mellon professor who teaches Game Design, recently gave a presentation at the Design Innovate Communicate Entertain (DICE) 2010 summit where he speaks about recent successes in game design in which games break into reality, drawing on ones' real social networks or achievement metrics that transcend individual games. Towards the end of the presentation, he imagines a world where one day we might earn bonus points simply by brushing our teeth or high-fiving friends. The video clip below shows the relevant segment and the full video can be found at G4TV.
So what about the life sciences? Where are the scientific bonus points? One idea proposed by Martijn Van Iersel, a developer for Pathvisio and WikiPathways, as a Google Summer of Code project is the development of a Scientific Karma website. The proposal calls for some standardized and automated way for contributions to various online communities to be scored. The website would display a list of wikis and communities that researchers have contributed to and a contributor's score for that site. Active contribution to online databases is important as it enhances all of our understandings and access to larger datasets helps research projects draw better conclusions about increasingly complex biological systems.
Bornmann, L., & Daniel, H. (2008). The state of h index research. Is the h index the ideal way to measure research performance? EMBO reports, 10 (1), 2-6 DOI: 10.1038/embor.2008.233
Friday, December 11, 2009
Google and Microsoft Sued by Mini Music Label
Blue Destiny Records has sued both Google and Microsoft for allegedly "facilitating and enabling" distribution of copyrighted songs, illegally. The suit alleges that RapidShare runs "a distribution center for unlawful copies of copyrighted works." RapidShare is helped by Google and Microsoft, which benefit from the ad relationships, according to the suit. Blue Destiny has attempted to links to pages with RapidShare links to their music via DMCA takedown notices, and Google has, apparently, not complied, while Microsoft's Bing site has removed the links. RapidShare for its part is based outside of the US and does not accept DMCA notices.
Wednesday, May 13, 2009
Lawsuit Filed Over Patenting Cancer Genes
 When Genae Girard attempted to get a second opinion about her risk for ovarian cancer she found out that Myriad Genetics had patented two genes BRCA1 and BRCA2 associated with cancer and there was no other tests. The ACLU has filed a lawsuit challenging the patent and Girard is joined by several others in the lawsuit: four other cancer patients, by professional organizations of pathologists with more than 100,000 members and by several individual pathologists and genetic researchers. Similar to the AACS encryption key controversy, Myriad Genetics has patented a specific piece of knowledge which is otherwise readily available, here is the DNA sequence for BRCA1. As Jan A. Nowak, president of the Association for Molecular Pathology, says in article "You can’t patent my DNA, any more than you can patent my right arm, or patent my blood." The US National Institutes of Health reported that some 20 percent of the genome, amounting to thousands of genes, has been included in patents claims. For those of you keeping track, there are only about 25,000 genes in the human genome.
When Genae Girard attempted to get a second opinion about her risk for ovarian cancer she found out that Myriad Genetics had patented two genes BRCA1 and BRCA2 associated with cancer and there was no other tests. The ACLU has filed a lawsuit challenging the patent and Girard is joined by several others in the lawsuit: four other cancer patients, by professional organizations of pathologists with more than 100,000 members and by several individual pathologists and genetic researchers. Similar to the AACS encryption key controversy, Myriad Genetics has patented a specific piece of knowledge which is otherwise readily available, here is the DNA sequence for BRCA1. As Jan A. Nowak, president of the Association for Molecular Pathology, says in article "You can’t patent my DNA, any more than you can patent my right arm, or patent my blood." The US National Institutes of Health reported that some 20 percent of the genome, amounting to thousands of genes, has been included in patents claims. For those of you keeping track, there are only about 25,000 genes in the human genome. Sunday, March 01, 2009
Marine One Blueprints Found on P2P
A P2P monitoring company Tiversa found "a file containing entire blueprints and avionics package for Marine One, which is the president's helicopter," on a P2P network. The original computer was traced back to a defense contractor in Bethesda, MD. Retired Gen. Wesley Clark, an adviser to Tiversa, said, "We found where this information came from. We know exactly what computer it came from. I'm sure that person is embarrassed and may even lose their job, but we know where it came from and we know where it went."
Sunday, February 22, 2009
Google Scholar to CiteULike
A Greasemonkey script that places a CiteULike link next to article links in Google Scholar; the following sites are supported, currently. You may have to look at "All X versions" to see the CiteULike link. For life sciences, the keyword "ncbi" or "pubmed" should make the right versions appear.
"http://www.ncbi.nlm.nih.gov/entrez/", // Entrez PubMed "http://www.ncbi.nlm.nih.gov/pubmed/", // PubMed "http://www.pubmedcentral.nih.gov/", // PubMed Central "http://highwire.stanford.edu/", // HighWire "http://scitation.aip.org/", // AIP Scitation "http://citeseer.ist.psu.edu/", // CiteSeer "http://arxiv.org/", // arXiv.org e-Print archive "http://www.worldcatlibraries.org/", // OCLC WorldCat "http://www.ingentaconnect.com/", // IngentaConnect "http://www.agu.org/", // American Geophysical Union "http://ams.allenpress.com/", // American Meteorological Society "http://www.anthrosource.net/", // Anthrosource "http://portal.acm.org/", // Association for Computing Machinery (ACM) portal "http://www.usenix.org/", // Usenix "http://bmj.bmjjournals.com/", // BMJ "http://ieeexplore.ieee.org/", // IEEE Xplore "http://journals.iop.org/", // IoP Electronic Journals "http://www.ams.org/mathscinet", // MathSciNet "http://www.nature.com/", // Nature "http://www.plosbiology.org/", // PLoS Biology "http://www.sciencemag.org/", // Science "http://www.kluweronline.com/", // now SpringerLink "http://links.jstor.org/", // JSTOR "http://www.springerlink.com/", // SpringerLink "http://www.metapress.com/", // MetaPress "http://www3.interscience.wiley.com/", // Wiley InterScience "http://www.sciencedirect.com/" // ScienceDirect
Sunday, July 27, 2008
Playstation Cell Processor Used For Plane Safety
 David Bader from Georgia Tech's College of Computing is researching uses for the Playstation 3 Cell chip for monitoring vibrations of plane components during flight that can act as a early warning detection system. The school has announced seven pilot projects through its Sony-Toshiba-IBM (STI) Center of Competence in areas such as seismic monitoring, electronic financial services, secure communications and biology research.
David Bader from Georgia Tech's College of Computing is researching uses for the Playstation 3 Cell chip for monitoring vibrations of plane components during flight that can act as a early warning detection system. The school has announced seven pilot projects through its Sony-Toshiba-IBM (STI) Center of Competence in areas such as seismic monitoring, electronic financial services, secure communications and biology research.
Thursday, May 15, 2008
The Design and Characterization of Two Proteins with 88% Sequence Identity But Different Structure
The protein studied was a cell wall protein from the Streptococcus bacteria called protein G. The protein is composed of two separate domains: the first domain (GA) has the ability to bind to human serum albumin (HSA) found in blood and the second one (GB) can bind to a region of IgG, a human antibody also found in blood. The two domains have different folds 3-alpha and alpha/beta folds, respectively. Each of the domains have positive free energies (delta G) indicating that the folded states are more stable. These two domains (GA and GB) originated as PSD1 and GB1, each containing 56 amino acids. In the GA domain, only 47 of the amino acids were structured; the other 9 were disordered.
In the image below, amino acids shown in blue are identities and those shown in red are nonidentities. Spheres indicate mutations introduced in each design cycle.

Altering GA within the structured 47 amino acids can change the equilibrium of states from the GA 3-alpha fold to the alpha/beta fold of the GB domain. The goal of the project was to investigate the number of amino acids that could be changed in each protein while retaining biological function.
The first step was to introduce both the IgG and the HSA-binding epitopes (the parts of a molecule that antibodies bind to) to both proteins. The HSA binding site in GA is composed of 7 amino acids, while the IgG binding site is composed of 4 amino acids in the central helix, one amino acid in the beta-3 strand and the main chain contacts. The latent IgG binding site was introduced to GA through 3 mutations, while the HSA binding site was introduced into GB with 5 mutations; the mutants were denoted as GA30 and GB30, respectively, as denoting their 30% similarity.
Several methods were used to characterize the proteins. The secondary structures of both proteins were determined using circular dichroism (CD), a form of spectroscopy looking at differential absorption of polarized light [3]. Other methods used included: thermal denaturation to determine conformational stability, gel filtration to ensure monomeric behavior, and affinity chromatography to determine binding affinities to IgG and HSA. The addition of the latent binding sites did not produce any significant alterations in the thermodynamics of the unfolding reaction for each protein. The GB30 protein was less stable than the original, with a delta-G 3 kcal/mol lower. There was no variation in the heat capacity of the protein, which is correlated to the solvent-exposed area, meaning the hydrophobic cores of the proteins were not disturbed. Affinity chromatography showed that both GA30 and GB30 bound to IgG and HSA in a similar fashion to GA and GB.
Next, changes were made in the remaining 39 non-identity residues using random mutagenesis and phage display. One method for performing random mutagenesis is to introduce Mn2+ or Mg2+ into the system, which causes mutagenic conditions resulting in random errors during DNA replication [4]. Phage display relies on the bacteriophages that encode proteins displayed on the surface of the phage, which can be used to select for functionality. These proteins then can be selected for using immobilized antigens; the DNA encoding the protein will be located within the phage [5]. Mutations to the remaining 39 residues were categorized into one of three categories: (i) mutations tolerated independent of mutations to other residues, (ii) additional mutations that must be made to tolerate a mutation, and (iii) mutations found to be rare.
Mutations were made to 19 residues in GA30 and 8 residues in GB30 bringing the pair to 77% similarity. These new proteins showed a decrease in stability, a decrease in the free energy of unfolding, but both retained a delta-G greater than 4 kcal/mol at 25 degrees C. The proteins did not show a decrease in binding affinity and both remained monomeric.
The image below shows the stability curves for GA and GB.
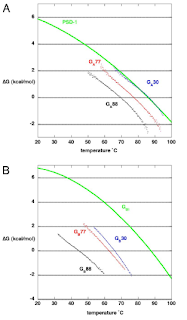
The proteins were brought to 88% similarity by changing two sites in GA77 and four sites in GB77. GA88 and GB88 are the result of 49 residues being the same: nine residues initially the same, 16 mutations in GA, 17 mutations in GB, and the addition of seven residues to GA. The CD spectrum remained close to that of the original proteins. The stability of both proteins further decreased to 4 kcal/mol for GA88 and 2 kcal/mol for GA88 at 25 degrees C. Both GA88 and GB88 retained their binding specificity to HSA and IgG, respectively, but GA88 had a lower binding affinity than GA77. Both proteins remained monomeric at this level of similarity.
Mutation of the seven unique residues can shift the fold type of the proteins from either 99.9% 3-alpha fold to the 97% alpha/beta fold. This study has implications for researchers working on computational protein folding prediction because proteins may be able to exist in one of many stable folded states as a result of mutations. Secondly, this study showed that few mutations can alter the function of a protein given the inclusion of latent binding epitopes, which may not affect the function of the native state.
References
[1] Davidson AR. A folding space odyssey. Proc Natl Acad Sci U S A. 2008 Feb 26;105(8):2759-60. Epub 2008 Feb 19.
[2] Alexander PA, He Y, Chen Y, Orban J, Bryan PN. The design and characterization of two proteins with 88% sequence identity but different structure and function. Proc Natl Acad Sci U S A 2007 Jul 17 104(29):11963-8
[3] "Circular dichroism." Wikipedia, The Free Encyclopedia. 11 Mar 2008, 19:51 UTC. Wikimedia Foundation, Inc. 16 Mar 2008 .
[4] Pritchard L, et al. A general model of error-prone PCR. J Theor Biol. 2005 Jun 21;234(4):497-509.
[5] Sidhu SS, Koide S. Phage display for engineering and analyzing protein interaction interfaces. Curr Opin Struct Biol. 2007 Aug;17(4):481-7. Epub 2007 Sep 17.
Alexander, P.A., He, Y., Chen, Y., Orban, J., Bryan, P.N. (2007). The design and characterization of two proteins with 88% sequence identity but different structure and function. Proceedings of the National Academy of Sciences, 104(29), 11963-11968. DOI: 10.1073/pnas.0700922104
Friday, May 02, 2008
Copy Number Variations in Human Disease
Several groups have turned their focus to understanding the role of amplifications or deletions, which characterize CNVs, in diseases such as mental disease and cancer. Over 50 regions were found to be associated possessing either unique amplifications or deletions in a cohort of 60 cancer patients [Lucito et al 2007]. The choice by Redon et al. to use the same samples that make up the HapMap dataset was to ease the integration of analysis comparison of CNV and SNP information that makes up the HapMap data.Correlation between CNV associations have been compared to known associations determined from SNP data. Sutrala et al. looked at 15 major candidate genes for schizophrenia including dysbindin (DTNBP1), neuregulin (NRG1), RGS4 and DISC1 and found no copy number variations at these loci. This is consistent with previous studies in the area, suggesting that the two types of variations act independently. This observation is further corroborated by a model of complex traits that showed that CNVs were able to account for 18% of gene expression variation, while SNPs could account for only 84% of the variation [Stranger 2007]. This study by Stranger et al. showed that the phenotypic variations explained by both types of structural variations were largely mutually exclusive in the HapMap samples of lymphoblastoid cell lines.
This new understanding about the potential importance of CNVs brings with it new challenges for the analysts due to the lack of genotyped CNV data and a lack of methods for analyzing CNV data at the genome-wide scale. Stranger et al., McCarroll, and Altshuler point to the fact that the low resolution of CNV data and potential issues with measurement precision could lead to analysis inaccuracies. In 2007, a paper by McCarroll and Altshuler stated that out of the 1,500 CNVs that have been identified researchers have only genotyped 70 at a quality necessary to carry out linkage disequilibrium studies. A recent paper by Ionita-Laza et al. presents a new method for family-based association tests (FBATs) that circumvents the need for genotypes and relies on intensity values. Family-based association tests are a key method used by geneticists in determining the association of genetic markers and disease because many of the issues with the selection of controls can be avoided by using data made up of family members. Ionita-Laza et al. extended the FBAT statistic in such a way that the average intensity within a family corresponds to Mendelian transmissions. Using their method, Ionita-Laza et al. were able to determine potential associations between the SNP rs2240832 on chromosome 7 in a known CNV region and asthma in a sample of 400 parent/child trios [Ionita-Laza 2008].
Another problem exists with the use of CNV data as a result of the procedure for collecting CNV data using array comparative genomic hybridization (array CGH). The method looks at the relative hybridization intensities of labeled probes of test and reference genomic DNA to determine raw intensity values [Pinkel and Albertson 2005]. Conrad et al. show that the frequency of CNV deletions increases as the the size of the deletions became smaller. This presents a problem with the current use of array CGH methods because of the low resolutions afforded by the method is approximately 5–10 Mb [Conrad 2007].
The direction of research seems directed toward the integration CNVs and SNPs for the purposes of understanding the role of variation in human disease. But fundamental issues of data resolution and quality must be answered before this type of integration can take place. This is a part of the much larger problem of classifying and integrating information of sequence and structure variation in genomes. These variations include SNPs and CNVs discussed here, but also inversions, translocations, and deletions of varying scales which, currently, must be identified using different methods. Lee et al points out that the small number of findings linking CNVs to phenotypic differences has a negative effect on the expanded use of higher-resolution array CGH for clinical purposes, which in turn, affects the accumulation of CNV data that may prove useful.
[1] Redon R, et al. Global variation in copy number in the human genome. Nature 2006 Nov 23 444(7118):444-54.
[2] Lucito R, et al. Copy-number variants in patients with a strong family history of pancreatic cancer. Cancer Biol Ther. 2007 Oct;6(10):1592-9. Epub 2007 Jul 12.
[3] Sutrala SR, et al. Gene copy number variation in schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2007 Dec 28 [Epub ahead of print].
[4] Stranger BE, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007 Feb 9;315(5813):848-53.
[5] McCarroll SA, Altshuler DM. Copy-number variation and association studies of human disease. Nat Genet. 2007 Jul;39(7 Suppl):S37-42.
[6] Ionita-Laza I. et al. On the analysis of copy-number variations in genome-wide association studies: a translation of the family-based association test. Genet Epidemiol. 2008 Apr;32(3):273-84.
[7] Pinkel D, Albertson DG. Comparative genomic hybridization. Annu Rev Genomics Hum Genet. 2005;6:331-54.
[8] Scherer SW. et al. Challenges and standards in integrating surveys of structural variation. Nat Genet. 2007 Jul;39(7 Suppl):S7-15.
[9] Conrad, D.F. et al. A high-resolution survey of deletion polymorphism in the human genome. Nat Genet. 2006 Jan;38(1):75-81. Epub 2005 Dec 4.
[10] Lee C, et al. Copy number variations and clinical cytogenetic diagnosis of constitutional disorders. Nat Genet. 2007 Jul;39(7 Suppl):S48-54.
Redon, R., Ishikawa, S., Fitch, K.R., Feuk, L., Perry, G.H., Andrews, T.D., Fiegler, H., Shapero, M.H., Carson, A.R., Chen, W., Cho, E.K., Dallaire, S., Freeman, J.L., González, J.R., Gratacòs, M., Huang, J., Kalaitzopoulos, D., Komura, D., MacDonald, J.R., Marshall, C.R., Mei, R., Montgomery, L., Nishimura, K., Okamura, K., Shen, F., Somerville, M.J., Tchinda, J., Valsesia, A., Woodwark, C., Yang, F., Zhang, J., Zerjal, T., Zhang, J., Armengol, L., Conrad, D.F., Estivill, X., Tyler-Smith, C., Carter, N.P., Aburatani, H., Lee, C., Jones, K.W., Scherer, S.W., Hurles, M.E. (2006). Global variation in copy number in the human genome. Nature, 444(7118), 444-454. DOI: 10.1038/nature05329
Monday, February 11, 2008
Calculation of Drag Force on a Wintry Boston Day
Presented here is a calculation of drag force caused by the wind on a cold and windy Boston day. The equations were taken from the Wikipedia page on drag coefficient, which has further explanations of each term. Pressure and wind velocity information was taken from Weather.com
Drag Force: Dry Air Density:
Dry Air Density:  Ideal Gas Law Constant for Dry Air:
Ideal Gas Law Constant for Dry Air:  Wind Velocity:
Wind Velocity:  Drag Coefficient of an Upright Person: Cd=1.3
Drag Coefficient of an Upright Person: Cd=1.3
Frontal Area of a Person: A=0.75 m2
Air Pressure: P=101,500 Pa
Temperature: T=276.15 K
The force (Fd) comes out to about 76 N or the weight of a cat on top of you while lying down.
Drag Force:
 Dry Air Density:
Dry Air Density:  Ideal Gas Law Constant for Dry Air:
Ideal Gas Law Constant for Dry Air:  Wind Velocity:
Wind Velocity:  Drag Coefficient of an Upright Person: Cd=1.3
Drag Coefficient of an Upright Person: Cd=1.3Frontal Area of a Person: A=0.75 m2
Air Pressure: P=101,500 Pa
Temperature: T=276.15 K
The force (Fd) comes out to about 76 N or the weight of a cat on top of you while lying down.
Sunday, February 03, 2008
Prevent Windows Disk Cleanup From Compressing Old Files
This registry hack prevents Windows Disk Cleanup Wizard from calculating the amount of space saved from compressing files on the drive. To open the registry editor, hit Windows Key and the R key at the same time. In the dialog that comes up, type in regedit and click OK.
Navigate to this registry entry.
Double-click the (Default) key and remove the value data, click OK and close the registry editor. The value of the entry just deleted is {B50F5260-0C21-11D2-AB56-00A0C9082678} should there ever be a need to reinstate the file compression.
Navigate to this registry entry.
HKEY_LOCAL_MACHINE\SOFTWARE\Microsoft\Windows\
CurrentVersion\Explorer\VolumeCaches\Compress Old FilesDouble-click the (Default) key and remove the value data, click OK and close the registry editor. The value of the entry just deleted is {B50F5260-0C21-11D2-AB56-00A0C9082678} should there ever be a need to reinstate the file compression.
Wednesday, January 30, 2008
Trends in Drug Discovery and Drug Targets
Yildirim et. al analyzed over 4,200 drug entries in the DrugBank database as a network between drugs and their targets. DrugBank contains detailed drug data such as chemical composition and prescribed uses, as well as information about the protein targets of the drug (i.e. sequence, structure and pathway). DrugBank has two major sets of drug entries: the first category is US Food and Drug Administration (FDA)-approved drugs, which accounts for approximately 1,200 drugs, and a second set composed of more than 3,000 "experimental" drugs under investigation. Out of the set of FDA-approved medications there are nearly 400 distinct human proteins being targeted. The inclusion of experimental drugs increased the number of drug targets to about 1,000.
Yildirim et al. created a bipartite graph made up of two distinct sets of nodes: drugs and the protein drug targets (shown below). In this type of network graph, drugs can have connections to drug targets, but never to another drug. Out of the 890 FDA-approved drugs, 788 were found to share a drug target with another drug. There analysis showed that several drugs had over 10 target proteins including propiomazine (Largon), promazine (Sparine), olanzapine (Zyprexa, Zydis), and ziprasidone (Geodon); all of these drugs are prescribed as anti-psychotics. The most-targeted proteins included the histamine receptor H1 (HRH1) and the muscarinic 1 cholinergic receptor (CHRM1) with about 50 drugs targeting each protein. Two networks were then created from the bipartite graph: a drug network with drugs sharing common targets being connected and a target-protein (TP) network was created that includes only drug targets and connections are drawn between two proteins, if a drug targets both. In the TP network, it was shown that 305 out of the 394 drug targets shared a connection.
 The giant component, the largest connected part of the network, for the drug network was 476 and 122 for the TP network, both smaller than would be expected using randomly generated networks with the same number of nodes and edges. This leads to the conclusion that highly connected nodes in the drug and TP networks are connected to other similarly connected nodes. By looking at the anti-psychotics with many target proteins listed above, we see this trend in that they all share a common function.
The giant component, the largest connected part of the network, for the drug network was 476 and 122 for the TP network, both smaller than would be expected using randomly generated networks with the same number of nodes and edges. This leads to the conclusion that highly connected nodes in the drug and TP networks are connected to other similarly connected nodes. By looking at the anti-psychotics with many target proteins listed above, we see this trend in that they all share a common function.From the experimental drug set, an additional 617 protein targets were identified from the approximately 800 experimental drugs with known targets. The giant component was determined to be 596 for the network of proteins targeted with the inclusion of experimental drugs. This is larger than would be expected for random networks, which was calculated as 551 (+/- 10). This differs from the result of using only the targets of FDA-approved drug targets, leading to the conclusion that experimental drug targets are more diversified than currently approved FDA-medications indicating a trend towards polypharmacology (drugs with multiple targets). A large segment, 62%, of FDA-approved drugs target proteins on cell membranes. This contrasts with many experimental drugs attempting to derive a treatment by targeting cellular components inside the cell; only about 40% of experimental drugs target membrane proteins. But looking at the drugs approved from 1996 to 2006, 69% of them target membrane proteins showing that while drug-discovery research is expanding targets, it has not been overly fruitful.
For the next part of their analysis, Yildirim et al. attempted to determine the essentiality of targets by identifying target proteins in a protein-protein interaction (PPI) map. They found that 262 targets were present and on average drug targets had more connections than other proteins in the network. Essential proteins are stated as proteins whose orthologous encoding genes in mice were necessary to produce a viable mice in gene knockout experiments; the quantitative measure is the number of connections of the essential predicted proteins.
 Using data from Online Mendelian Inheritance in Man (OMIM) Morbid Map, which contains disorder to disease gene associations, a human disease-gene (HDG) network was produced drawing connections between two genes which share a common disease. Only 166 out of 1,777 disease-related genes encode for drug targets; 43% are associated with multiple diseases. Including the target proteins of the experimental drugs, the percentage associated with multiple diseases dropped to 26%, which shows a trend to towards targeting disease genes more specifically.
Using data from Online Mendelian Inheritance in Man (OMIM) Morbid Map, which contains disorder to disease gene associations, a human disease-gene (HDG) network was produced drawing connections between two genes which share a common disease. Only 166 out of 1,777 disease-related genes encode for drug targets; 43% are associated with multiple diseases. Including the target proteins of the experimental drugs, the percentage associated with multiple diseases dropped to 26%, which shows a trend to towards targeting disease genes more specifically.The authors suggest that most drugs are palliative in nature (relieve symptoms, but do not address the specific genetic cause) due to the distance between the disease-gene and target proteins closely matching the distances for randomized pairings of drug target and disease-gene pairings (shown below). Since 1996, the fraction of disease-gene and drug-target with shorter distances has increased (also shown below). Diseases having drugs that are more directly targeted included cancer, endocrine, psychiatric, and respiratory disease classes. Developmental, muscular, and ophthalmologic diseases had longer distances from target protein. For medications treating advanced cases of cancer, the path from drug target to disease-gene is longer indicating that the drugs are more palliative.

 One thing lacking from this paper information of the efficacy of the drugs. Overington et al. stated in 2006 that the number of targeted genes would increase to over 600 genes, if you assume that drugs would have some effect on proteins showing 50% identity similarity. Another measure of efficacy that could have been used is binding-affinities to give strengths to the protein-protein interactions. Another interesting piece of information that Yildirim et al. do not highlight is the categories of target proteins. In the same 2006 perspectives paper by Overington et al., drug targets are classified by gene family and they show that over 50% of drugs fell into one of four categories: class I G-protein coupled receptors (GPCRs), nuclear receptors, ligand-gated ion channels, and voltage-gated ion channels.
One thing lacking from this paper information of the efficacy of the drugs. Overington et al. stated in 2006 that the number of targeted genes would increase to over 600 genes, if you assume that drugs would have some effect on proteins showing 50% identity similarity. Another measure of efficacy that could have been used is binding-affinities to give strengths to the protein-protein interactions. Another interesting piece of information that Yildirim et al. do not highlight is the categories of target proteins. In the same 2006 perspectives paper by Overington et al., drug targets are classified by gene family and they show that over 50% of drugs fell into one of four categories: class I G-protein coupled receptors (GPCRs), nuclear receptors, ligand-gated ion channels, and voltage-gated ion channels.Yildirim, M.A., Goh, K., Cusick, M.E., Barabasi, A., Vidal, M. (2007). Drug-target network. Nature Biotechnology, 25(10), 1119-1126. DOI: 10.1038/nbt1338
Overington, J.P, et al. How many drug targets are there? Nature Reviews Drug Discovery, Vol. 5, No. 12., pp. 993-996.
Thursday, December 27, 2007
The Genomics Revolution and the State of Personalized Medicine
 The seeds of the Human Genome Project (HGP) were laid during the 1984 Alta Summit in the Wasatch Mountains of Utah. Scientists gathered as part of a meeting sponsored by the Department of Energy (DOE) and the International Commission for Protection Against Environmental Mutagens and Carcinogens. They came with the specific question: "Could new methods permit direct detection of mutations, and more specifically could any increase in the mutation rate among survivors of the Hiroshima and Nagasaki bombings be detected (in them or in their children)?" [1] Seventeen years after that summit the first publications on the preliminary analyses on the working drafts human genome were published by Nature and Science journals in February 2001 by two competing groups: the International Human Genome Mapping Consortium and the Celera Genomics Sequencing Team. [2-3] The same issue of Nature featured a perspective on the potential importance to medicine of the 1.4 million single nucleotide polymorphisms (SNPs) mapped in the genome [4]. One pragmatic approach in interpreting the success of the HGP is in determining its current impact on medicine.
The seeds of the Human Genome Project (HGP) were laid during the 1984 Alta Summit in the Wasatch Mountains of Utah. Scientists gathered as part of a meeting sponsored by the Department of Energy (DOE) and the International Commission for Protection Against Environmental Mutagens and Carcinogens. They came with the specific question: "Could new methods permit direct detection of mutations, and more specifically could any increase in the mutation rate among survivors of the Hiroshima and Nagasaki bombings be detected (in them or in their children)?" [1] Seventeen years after that summit the first publications on the preliminary analyses on the working drafts human genome were published by Nature and Science journals in February 2001 by two competing groups: the International Human Genome Mapping Consortium and the Celera Genomics Sequencing Team. [2-3] The same issue of Nature featured a perspective on the potential importance to medicine of the 1.4 million single nucleotide polymorphisms (SNPs) mapped in the genome [4]. One pragmatic approach in interpreting the success of the HGP is in determining its current impact on medicine.Overall, the project took 13 years to complete in 2003; whereby basic science has benefited from the HGP [5]. Research in surrounding fields has progressed at an incredible rate creating a veritable alphabet soup of -omics-related projects: peptidomics, proteomics, genomics, lipidomics, transcriptomics, metabolomics, metallomics, glycomics, interactomics, spliceomics, and ORFeomics. Researchers who worked on the original effort to sequence the genome turned their efforts to sequencing the genome of one individual since the HGP dealt with a conglomeration of DNA from several individuals. Personalized genomes are able to provide more relevant information about specific conditions in individuals. J. Craig Venter was the first individual for which a diploid genome sequence was characterized in September 2007 [6]. In the publication on Venter's genome, a table is provided containing many markers for common diseases and traits showing the potential for self-examination possible from knowing our genetic sequence. For instance, the list revealed that Venter is more likely to have wet earwax due to the sequence of his ABCC11 gene [6]. It also reveals that Venter is at a higher risk of Alzhimer's disease; information, which may be relevant to health-practitioners diagnosing aliments.
Unfortunately, much of the genomics revolution in science has side-stepped health-practitioners in part due to cost. Venter's genome cost about $70 million to sequence [7]. His genome was followed shortly thereafter by the genome of James Watson employing the sequencing technology of 454 Life Sciences which cost less than $1 million dollars [8]. Estimated costs for a sequencing an individual's genome is approximately $100,000 using the most current technology [7]. The X-Prize Foundation has set up an a genomics X-Prize worth $10 million for the first team able to sequence the genomes of 100 individuals in 10 days; any technologies borne from this accomplishment would almost certainly drive costs down further [8].
There is tension between those geneticists that feel that publicity gained from the sequencing of the genomes of the wealthy and celebrities will garner more attention and support from the public and those who feel that it a misuse of genomics. A news article in the May 2007 issue of Nature shows this unease; Francis Collins, director of the National Human Genome Research Institute (NHGRI), was quoted as saying that sequencing of scientists with "strong financial positions ... [is] contrary to what the genome project aimed to achieve." The NHGRI plans to sequence 100 individual genomes in the next several years and wants to maximize the information obtained from these individuals. Ideas for candidates include those individuals with rare genetic disorders and individuals who have already been studied as part of the International HapMap Project, which published its analyses in the October 2007 [8-9].
Genomics in clinical settings is not routinely utilized, but it is an area that deserves increasing focus at medical schools and continuing education programs. Essential knowledge for current health-practitioners includes understanding available genetic tests for common diseases in pre-symptomatic patients and their uses in diagnoses of symptomatic patients. Our current understanding of the interactions between environmental and genetic factors and the small role that any individual genetic marker may make to complex human diseases undercuts the possibilities for determining more from genetic tests [10]. In a survey study conducted by Finn et al. of US and Canadian psychiatrists, it was shown that fewer than 25% of those surveyed felt prepared or competent to discuss information from genetic analyses with patients and their families [11]. Furthermore, the attitudes of many health-practitioners can be summarized as the HGP as being interesting and potentially useful one day, but having no dramatic effects on health-care in the near future, which has left many wondering "What will genetics and genomics do for me now, and how will they improve patient outcomes?" [12]. Guttenmacher proposes several recommendations to bridge the gap between the basic sciences and medical training, including increased focus on the relationship between common diseases (as opposed to rare Mendelian diseases) and genetics so that health-practitioners begin to think about the genetic factors contributing to the expression of diseases in patients [12].
Science tends to work in leaps and bounds, and while the cost has been prohibitive up to this point, two companies have recently announced new genotyping services for about $1,000. On November 17, 2007, the New York Times contained an article about two new companies; deCODE Genetics, an Icelandic company, will offer to genotype 1 million SNPs from cheek scrapings for $985, and a Google-financed company, 23andMe, will soon announce a similar service that will test 650,000 SNPs [13]. But given the caution with which trained genetic counselors approach genomic information one wonders the use of this data to the average person and the misinformation that might arise from misreading.
[1] Robert Cook-Deegan. The Alta Summit, December 1984. Genomics 5. 661-663 (1989)
[2] The International Human Genome Mapping Consortium. A physical map of the human genome. Nature 409, 934-941 (2001).
[3] The Celera Genomics Sequencing Team. The sequence of the human genome. Science. 291(5507):1304-51. (2001).
[4] Chakravarti, A. Single nucleotide polymorphisms: . . .to a future of genetic medicine. Nature. 409(6822):822-3 (2001).
[5] Francis S. Collins, Michael Morgan, Aristides Patrinos. The Human Genome Project: Lessons from Large-Scale Biology. Science. 300(5617):286-90 (2003).
[6] Levy, S. et al. The Diploid Genome Sequence of an Individual Human. PLoS Biol. 5(10):e254 (2007).
[7] May, M. Venter Sequenced. Nature. Vol 25 10. 1071 (2007).
[8] Check, E. Celebrity genomes alarm researchers. Nature 447, 358-359 (2007).
[9] The International SNP Map Working Group. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 409(6822):928-33 (2001).
[10] McGuire. A.L. Medicine. The Future of Personal Genomics. Science. 317(5845):1687 (2007).
[11] Finn, C. T. et al. Psychiatric genetics: a survey of psychiatrists’ knowledge, attitudes, opinions, and practice patterns. J. Clin. Psychiatry 66, 821–830 (2005).
[12] Guttmacher AE, Porteous ME, McInerney JD. Educating health-care professionals about genetics and genomics. Nat Rev Genet. 8(2):151-7 (2007).
[13] Wade, N. Experts Advise a Grain of Salt With Mail-Order Genomes, at $1,000 a Pop. New York Times. 17 Nov. 2007. Accessed: http://www.nytimes.com/2007/11/17/us/17genome.html
Monday, December 03, 2007
List of Facebook Beacon Partners
 This is a partial list of the participating websites that have formed partnerships with Facebook Beacon. The list was obtained from Facebooking101.com An article by Juan Carlos Perez at PC World indicates that participating websites may be sending data about users to Facebook even they are logged out and have declined to broadcast their activities to Facebook friends. MoveOn.org has begun a petition stating that Facebook must respect user's online privacy and information should only be used with explicit permission. Additionally, Freakin Beacon, a Firefox extension, has been created which will display a blue icon in the status bar whenever a user visits a Beacon site.
This is a partial list of the participating websites that have formed partnerships with Facebook Beacon. The list was obtained from Facebooking101.com An article by Juan Carlos Perez at PC World indicates that participating websites may be sending data about users to Facebook even they are logged out and have declined to broadcast their activities to Facebook friends. MoveOn.org has begun a petition stating that Facebook must respect user's online privacy and information should only be used with explicit permission. Additionally, Freakin Beacon, a Firefox extension, has been created which will display a blue icon in the status bar whenever a user visits a Beacon site.- AllPosters.com
- Blockbuster
- Bluefly.com
- Busted Tees
- CBS Interactive (CBSSports.com & Dotspotter)
- Citysearch
- CollegeHumor
- eBay
- echomusic
- ExpoTV
- Fandango
- Gamefly
- Hotwire
- iWon
- Joost
- Kiva
- Kongregate
- LiveJournal
- Live Nation
- Mercantila
- National Basketball Association
- NYTimes.com
- Overstock.com
- Pronto.com
- (RED)
- Redlight
- SeamlessWeb
- Sony Online Entertainment LLC
- Sony Pictures
- STA Travel
- The Knot
- TripAdvisor
- Travelocity
- Travel Ticker
- TypePad
- viagogo
- Vox
- Yelp
- WeddingChannel.com
- Zappos.com
Tuesday, October 23, 2007
Using Gmail IMAP with Windows Mobile
Gmail recently introduced IMAP for Gmail on select accounts. Windows Mobile phones can be configured to take advantage of the new IMAP feature using the following instructions.
Windows Mobile phones can be configured to take advantage of the new IMAP feature using the following instructions.
 Windows Mobile phones can be configured to take advantage of the new IMAP feature using the following instructions.
Windows Mobile phones can be configured to take advantage of the new IMAP feature using the following instructions.- Open the Messaging application
- Choose any of the accounts currently listed
- Click "Menu" then "Options"
- Select "New Account"
- On the "E-mail Setup" screen, fill out your name and your email address, then uncheck "Attempt ..." and click next.
- Enter your username and password; leave "Domain" blank, and click "Next".
- For "Server type" and "Account name", enter IMAP4, then click "Next"
- For "Incoming server", enter: imap.gmail.com
- Check "Require SSL connection"
- For "Outgoing server", enter: smtp.gmail.com
- Check "Outgoing server requires authentication"
- Click "Next"
- Choose the number of previous days and amount of message to download, then click "Next".
- Choose how often Gmail will be checked, then click "Next".
- Setup in finished, click "Finish" to start downloading your Gmail messages.



Saturday, April 28, 2007
GMail Server Error - Temporary Error (502)
While attempting delete 120,000+ emails (730+ MB) from a GMail account I received the following error:

We’re sorry, but your Gmail account is currently experiencing errors. You won’t be able to log in while these errors last, but don’t worry, your account data and messages are safe. Our engineers are working to resolve this issue.
Please try logging in to your account again in a few minutes.

Monday, February 12, 2007
Older Americans and Complementary/Alternative Medicine
 Individuals aged 50 and older tend to have a higher rate of complementary and alternative medicine (CAM) use, but tend not to discuss CAM use with their physicians. NCCAM conducted a phone survey of 1,559 people aged 50 and older showed that 30% did not discuss CAM use because they did not they should. Women more than men tended to discuss CAM use, and individuals making $75,000 or more were 6% more likely to talk to their physicians about CAM use than people with lower incomes.
Individuals aged 50 and older tend to have a higher rate of complementary and alternative medicine (CAM) use, but tend not to discuss CAM use with their physicians. NCCAM conducted a phone survey of 1,559 people aged 50 and older showed that 30% did not discuss CAM use because they did not they should. Women more than men tended to discuss CAM use, and individuals making $75,000 or more were 6% more likely to talk to their physicians about CAM use than people with lower incomes.There is the risk of drug interactions with some herbal remedies. The Food and Drug Administration (FDA) released a Public Health Advisory citing a paper in the (Journal of the American Medical Association) showing drug interactions between indinavir, a protease inhibitor used to treat HIV infections and St. John's wort. St. John's wort is often recommended in treating depression and anxiety.
Subscribe to:
Posts (Atom)










