The protein studied was a cell wall protein from the Streptococcus bacteria called protein G. The protein is composed of two separate domains: the first domain (GA) has the ability to bind to human serum albumin (HSA) found in blood and the second one (GB) can bind to a region of IgG, a human antibody also found in blood. The two domains have different folds 3-alpha and alpha/beta folds, respectively. Each of the domains have positive free energies (delta G) indicating that the folded states are more stable. These two domains (GA and GB) originated as PSD1 and GB1, each containing 56 amino acids. In the GA domain, only 47 of the amino acids were structured; the other 9 were disordered.
In the image below, amino acids shown in blue are identities and those shown in red are nonidentities. Spheres indicate mutations introduced in each design cycle.

Altering GA within the structured 47 amino acids can change the equilibrium of states from the GA 3-alpha fold to the alpha/beta fold of the GB domain. The goal of the project was to investigate the number of amino acids that could be changed in each protein while retaining biological function.
The first step was to introduce both the IgG and the HSA-binding epitopes (the parts of a molecule that antibodies bind to) to both proteins. The HSA binding site in GA is composed of 7 amino acids, while the IgG binding site is composed of 4 amino acids in the central helix, one amino acid in the beta-3 strand and the main chain contacts. The latent IgG binding site was introduced to GA through 3 mutations, while the HSA binding site was introduced into GB with 5 mutations; the mutants were denoted as GA30 and GB30, respectively, as denoting their 30% similarity.
Several methods were used to characterize the proteins. The secondary structures of both proteins were determined using circular dichroism (CD), a form of spectroscopy looking at differential absorption of polarized light [3]. Other methods used included: thermal denaturation to determine conformational stability, gel filtration to ensure monomeric behavior, and affinity chromatography to determine binding affinities to IgG and HSA. The addition of the latent binding sites did not produce any significant alterations in the thermodynamics of the unfolding reaction for each protein. The GB30 protein was less stable than the original, with a delta-G 3 kcal/mol lower. There was no variation in the heat capacity of the protein, which is correlated to the solvent-exposed area, meaning the hydrophobic cores of the proteins were not disturbed. Affinity chromatography showed that both GA30 and GB30 bound to IgG and HSA in a similar fashion to GA and GB.
Next, changes were made in the remaining 39 non-identity residues using random mutagenesis and phage display. One method for performing random mutagenesis is to introduce Mn2+ or Mg2+ into the system, which causes mutagenic conditions resulting in random errors during DNA replication [4]. Phage display relies on the bacteriophages that encode proteins displayed on the surface of the phage, which can be used to select for functionality. These proteins then can be selected for using immobilized antigens; the DNA encoding the protein will be located within the phage [5]. Mutations to the remaining 39 residues were categorized into one of three categories: (i) mutations tolerated independent of mutations to other residues, (ii) additional mutations that must be made to tolerate a mutation, and (iii) mutations found to be rare.
Mutations were made to 19 residues in GA30 and 8 residues in GB30 bringing the pair to 77% similarity. These new proteins showed a decrease in stability, a decrease in the free energy of unfolding, but both retained a delta-G greater than 4 kcal/mol at 25 degrees C. The proteins did not show a decrease in binding affinity and both remained monomeric.
The image below shows the stability curves for GA and GB.
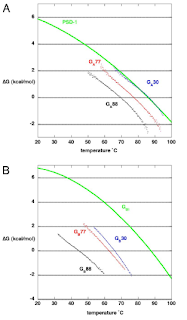
The proteins were brought to 88% similarity by changing two sites in GA77 and four sites in GB77. GA88 and GB88 are the result of 49 residues being the same: nine residues initially the same, 16 mutations in GA, 17 mutations in GB, and the addition of seven residues to GA. The CD spectrum remained close to that of the original proteins. The stability of both proteins further decreased to 4 kcal/mol for GA88 and 2 kcal/mol for GA88 at 25 degrees C. Both GA88 and GB88 retained their binding specificity to HSA and IgG, respectively, but GA88 had a lower binding affinity than GA77. Both proteins remained monomeric at this level of similarity.
Mutation of the seven unique residues can shift the fold type of the proteins from either 99.9% 3-alpha fold to the 97% alpha/beta fold. This study has implications for researchers working on computational protein folding prediction because proteins may be able to exist in one of many stable folded states as a result of mutations. Secondly, this study showed that few mutations can alter the function of a protein given the inclusion of latent binding epitopes, which may not affect the function of the native state.
References
[1] Davidson AR. A folding space odyssey. Proc Natl Acad Sci U S A. 2008 Feb 26;105(8):2759-60. Epub 2008 Feb 19.
[2] Alexander PA, He Y, Chen Y, Orban J, Bryan PN. The design and characterization of two proteins with 88% sequence identity but different structure and function. Proc Natl Acad Sci U S A 2007 Jul 17 104(29):11963-8
[3] "Circular dichroism." Wikipedia, The Free Encyclopedia. 11 Mar 2008, 19:51 UTC. Wikimedia Foundation, Inc. 16 Mar 2008 .
[4] Pritchard L, et al. A general model of error-prone PCR. J Theor Biol. 2005 Jun 21;234(4):497-509.
[5] Sidhu SS, Koide S. Phage display for engineering and analyzing protein interaction interfaces. Curr Opin Struct Biol. 2007 Aug;17(4):481-7. Epub 2007 Sep 17.
Alexander, P.A., He, Y., Chen, Y., Orban, J., Bryan, P.N. (2007). The design and characterization of two proteins with 88% sequence identity but different structure and function. Proceedings of the National Academy of Sciences, 104(29), 11963-11968. DOI: 10.1073/pnas.0700922104
No comments:
Post a Comment